Assessing Read Quality, Trimming and Filtering
Overview
Teaching: 45 min
Exercises: 45 minQuestions
How can I describe the quality of my data?
How can we get rid of sequence data that doesn’t meet our quality standards?
How do these methods differ when looking at Nanopore data?
Objectives
Interpret a FastQC plot summarizing per-base quality across all reads.
Interpret the NanoPlot output summarizing a Nanopore sequencing run
Filter Nanopore reads based on quality using the command line tool SeqKit
Quality control
Before assembling our metagenome from the the short-read Illumina sequences and the long-read Nanopore sequences, we need to apply quality control to both. The two types of sequence data require different QC methods. We will use:
- FastQC to examine the quality of the short-read Illumina data
- NanoPlot to examine the quality of the long-read Nanopore data and Seqkit to trim and filter them.
Illumina Quality control using FastQC
Reminder of the FASTQ format
In the FASTQ file format, each ‘read’ (i.e. sequence) is composed of four lines:
Line Description 1 Always begins with ‘@’ and gives the sequence identifier and an optional description 2 The actual DNA sequence 3 Always begins with a ‘+’ and sometimes the same info in line 1 4 Has a string of characters which represent the PHRED quality score for each of the bases in line 2; must have same number of characters as line 2
We can examine the first read in the FASTQ file using head to print the first four lines.
Move into the directory containing the illumina FASTQ files using cd:
cd cs_course/data/illumina_fastq/
Now use head to view the first 4 lines of the ERR2935805.fastq file
head -n 4 ERR2935805.fastq
@ERR2935805.1 HWI-C00124:284:HWTT2BCXY:1:1101:1247:2214 length=202
GATGGCGATAGAAGTCAAGTCTTTATTTTATGAAACCGCCATCATTAGTAGTATTTTATTTGGGCTCCCTTTTATAGGGACGGATATTTATGAGAATCAGCNAAAAAATCTACNCCTTCCTGAAANNANNAACNNNCAGGGTCTGACGATTTTCCTGCTGGGGTGGGAAATTGCCAGATAAAACAATATTGTGATTATCTCT
+ERR2935805.1 HWI-C00124:284:HWTT2BCXY:1:1101:1247:2214 length=202
GAGGGGIIIIIIGIIGIGIIGIGIGIIIIGIIGGGIGGGGGGGIIGIIIIIIIGGIGGIIIIGGGGGGIIGIIIGGGIIGGGGAGGGAGGGIAGGGGGGGA#<GGAIIIIIGI#<<GGGIGGGGA##<##<<G###<<GAGGIGGIIIIIIIIIGGIIIIIIIIIIGIIGGGGGAGGGGIIIGGIIGIGIGIGGIGGIGGG.
The quality score of this read is on line 4.
GAGGGGIIIIIIGIIGIGIIGIGIGIIIIGIIGGGIGGGGGGGIIGIIIIIIIGGIGGIIIIGGGGGGIIGIIIGGGIIGGGGAGGGAGGGIAGGGGGGGA#<GGAIIIIIGI#<<GGGIGGGGA##<##<<G###<<GAGGIGGIIIIIIIIIGGIIIIIIIIIIGIIGGGGGAGGGGIIIGGIIGIGIGIGGIGGIGGG.
PHRED score reminder
Quality encoding: !"#$%&'()*+,-./0123456789:;<=>?@ABCDEFGHIJ | | | | | Quality score: 01........11........21........31........41Quality is interpreted as the probability of an incorrect base call. To make it possible to line up each individual nucleotide with its quality score, the numerical score is encoded by a single character. The quality score represents the probability that the corresponding nucleotide call is incorrect. It is a logarithmic scale so a quality score of 10 reflects a base call accuracy of 90%, but a quality score of 20 reflects a base call accuracy of 99%.
The PHRED quality scores for the majority of the bases in this read are between 31-41.
Rather than assessing every read in the raw data by hand we can use FastQC to visualise the quality of the whole sequencing file.
First, we are going to organise our analysis by creating a directory to contain the output of all of the analysis we generate in this course.
The mkdir command can be used to make a new directory. Using the -p flag for mkdir allows it to create a new directory, even if one of the parent directories doesn’t already exist. It also suppresses errors if the directory already exists, without overwriting that directory.
Return to your home directory (/home/csuser)
cd
Create the directories analysis/qc/illumina_qc inside cs_course
mkdir -p cs_course/analysis/qc/illumina_qc
You might want to use ls to check those nested directories have been made.
Now we have created the directories we are ready to start the quality control of the Illumina data.
FastQC has been installed on your instance and we can run it with the -h flag to display the help documentation and remind ourselves how to use it and all the parameters available:
$ fastqc -h
FastQC seq help documentation
FastQC - A high throughput sequence QC analysis tool SYNOPSIS fastqc seqfile1 seqfile2 .. seqfileN fastqc [-o output dir] [--(no)extract] [-f fastq|bam|sam] [-c contaminant file] seqfile1 .. seqfileN DESCRIPTION FastQC reads a set of sequence files and produces from each one a quality control report consisting of a number of different modules, each one of which will help to identify a different potential type of problem in your data. If no files to process are specified on the command line then the program will start as an interactive graphical application. If files are provided on the command line then the program will run with no user interaction required. In this mode it is suitable for inclusion into a standardised analysis pipeline. The options for the program as as follows: -h --help Print this help file and exit -v --version Print the version of the program and exit -o --outdir Create all output files in the specified output directory. Please note that this directory must exist as the program will not create it. If this option is not set then the output file for each sequence file is created in the same directory as the sequence file which was processed. --casava Files come from raw casava output. Files in the same sample group (differing only by the group number) will be analysed as a set rather than individually. Sequences with the filter flag set in the header will be excluded from the analysis. Files must have the same names given to them by casava (including being gzipped and ending with .gz) otherwise they won't be grouped together correctly. --nano Files come from nanopore sequences and are in fast5 format. In this mode you can pass in directories to process and the program will take in all fast5 files within those directories and produce a single output file from the sequences found in all files. --nofilter If running with --casava then don't remove read flagged by casava as poor quality when performing the QC analysis. --extract If set then the zipped output file will be uncompressed in the same directory after it has been created. By default this option will be set if fastqc is run in non-interactive mode. -j --java Provides the full path to the java binary you want to use to launch fastqc. If not supplied then java is assumed to be in your path. --noextract Do not uncompress the output file after creating it. You should set this option if you do not wish to uncompress the output when running in non-interactive mode. --nogroup Disable grouping of bases for reads >50bp. All reports will show data for every base in the read. WARNING: Using this option will cause fastqc to crash and burn if you use it on really long reads, and your plots may end up a ridiculous size. You have been warned! --min_length Sets an artificial lower limit on the length of the sequence to be shown in the report. As long as you set this to a value greater or equal to your longest read length then this will be the sequence length used to create your read groups. This can be useful for making directly comaparable statistics from datasets with somewhat variable read lengths. -f --format Bypasses the normal sequence file format detection and forces the program to use the specified format. Valid formats are bam,sam,bam_mapped,sam_mapped and fastq -t --threads Specifies the number of files which can be processed simultaneously. Each thread will be allocated 250MB of memory so you shouldn't run more threads than your available memory will cope with, and not more than 6 threads on a 32 bit machine -c Specifies a non-default file which contains the list of --contaminants contaminants to screen overrepresented sequences against. The file must contain sets of named contaminants in the form name[tab]sequence. Lines prefixed with a hash will be ignored. -a Specifies a non-default file which contains the list of --adapters adapter sequences which will be explicity searched against the library. The file must contain sets of named adapters in the form name[tab]sequence. Lines prefixed with a hash will be ignored. -l Specifies a non-default file which contains a set of criteria --limits which will be used to determine the warn/error limits for the various modules. This file can also be used to selectively remove some modules from the output all together. The format needs to mirror the default limits.txt file found in the Configuration folder. -k --kmers Specifies the length of Kmer to look for in the Kmer content module. Specified Kmer length must be between 2 and 10. Default length is 7 if not specified. -q --quiet Supress all progress messages on stdout and only report errors. -d --dir Selects a directory to be used for temporary files written when generating report images. Defaults to system temp directory if not specified. BUGS Any bugs in fastqc should be reported either to simon.andrews@babraham.ac.uk or in www.bioinformatics.babraham.ac.uk/bugzilla/
We need to use just an input file and the -o flag to give the output directory: fastqc _inputfile_ -o _outputdirectory_
Navigate to your qc/ directory,
cd ~/cs_course/analysis/qc/
As we are using only one FASTQ file we can specify fastqc and then the location of the FASTQ file we want to analyse, and the illumina_qc output directory:
fastqc ~/cs_course/data/illumina_fastq/ERR2935805.fastq -o illumina_qc/
Press enter and you will see an automatically updating output message telling you the progress of the analysis. It should start like this:
Started analysis of ERR2935805.fastq
Approx 5% complete for ERR2935805.fastq
Approx 10% complete for ERR2935805.fastq
Approx 15% complete for ERR2935805.fastq
Approx 20% complete for ERR2935805.fastq
Approx 25% complete for ERR2935805.fastq
Approx 30% complete for ERR2935805.fastq
In total, it should take around ten minutes for FastQC to run on this fastq file (however, this depends on the size and number of files you give it). When the analysis completes, your prompt will return. So your screen will look something like this:
Approx 75% complete for ERR2935805.fastq
Approx 80% complete for ERR2935805.fastq
Approx 85% complete for ERR2935805.fastq
Approx 90% complete for ERR2935805.fastq
Approx 95% complete for ERR2935805.fastq
Analysis complete for ERR2935805.fastq
$
The FastQC program has created two new files within our
analysis/illumina_qc/ directory. We can see them by listing the contents of the illumina_qc folder
ls illumina_qc/
ERR2935805_fastqc.html ERR2935805_fastqc.zip
For each input FASTQ file, FastQC has created a .zip file and a .html file. The .zip file extension indicates that this is actually a compressed set of multiple output files. A summary report for our data is in the he .html file.
You need to transfer ERR2935805_fastqc.html from your AWS instance to your local computer to view it with a web browser.
To do this we will use the scp (secure copy protocol) command. You need to start a second terminal window that is not logged into the cloud instance and ensure you are in your cloudspan directory. This is important because it contains your pem file which will allow the scp command access to your AWS instance to copy the file.
Starting a new new terminal
Open your file manager and navigate to the
cloudspanfolder which should contain the login key file- Open your machine’s command line interface: Windows users: Right click anywhere inside the blank space of the file manager, then select Git Bash Here. Mac users: Open Terminal and type
cdfollowed by the absolute path that leads to yourcloudspanfolder. Press enter.- Check that you are in the right folder using
pwd
Now use scp to download the file.
The command will look something like:
scp -i login-key-instanceNNN.pem csuser@instanceNNN.cloud-span.aws.york.ac.uk:~/cs_course/analysis/qc/illumina_qc/ERR2935805_fastqc.html .
Remember to replace NNN with your instance number.
As the file is downloading you will see an output like:
scp -i login-key-instanceNNN.pem csuser@instanceNNN.cloud-span.aws.york.ac.uk:~/cs_course/analysis/qc/illumina_qc/ERR2935805_fastqc.html .
ERR2935805_fastqc.html 100% 591KB 1.8MB/s 00:00
Once the file has downloaded File Explorer (Windows) or Finder (Mac) to find the file and open it - it should open up in your browser.
Help!
If you had trouble downloading and viewing the file you can view it here: ERR2935805_fastqc.html
First we will look at the “Per base sequence quality” graph.
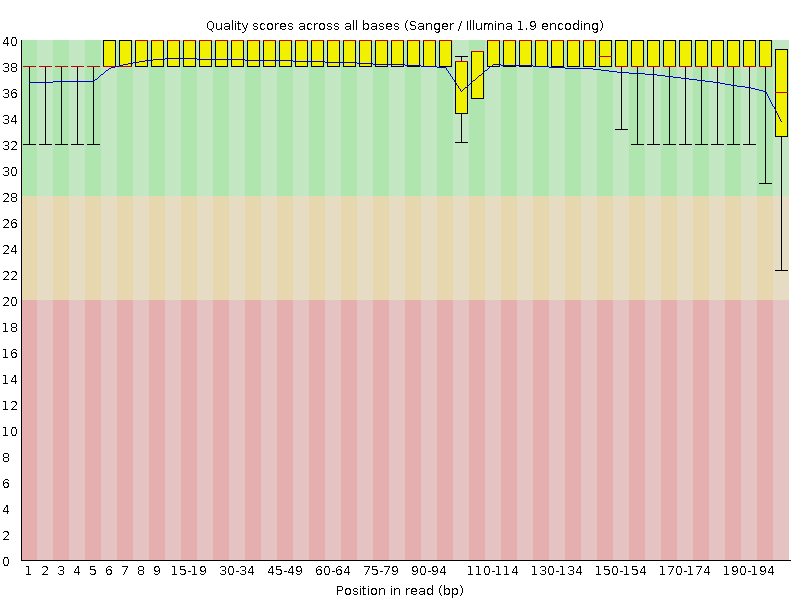
The x-axis displays the base position in the read, and the y-axis shows quality scores. In this example, the sample contains reads that are 202 bp long.
Each position has a box-and-whisker plot showing the distribution of quality scores for all reads at that position.
- The horizontal red line indicates the median quality score.
- The yellow box shows the 1st to 3rd quartile range (this means that 50% of reads have a quality score that falls within the range of the yellow box at that position).
- The whiskers show the absolute range, which covers the lowest (0th quartile) to highest (4th quartile) values.
The plot background is also color-coded to identify good (green), acceptable (yellow), and bad (red) quality scores.
In this sample, the quality values do not drop much lower than 32 at any position. This is a high quality score meaning the sequence is high quality. This means that we do not need to do any filtering. Lucky us!
We should also have a look at the “Adapter Content” graph which will show us where adapter sequences occur in the reads. Adapter sequences are short sequences that are added to the sample to aid during the preparation of the DNA library. They therefore don’t tell us anything biologically important and should be removed if they are present in high numbers. They might also be removed in the case of certain applications, such as ones when the base sequence needs to be particularly accurate.
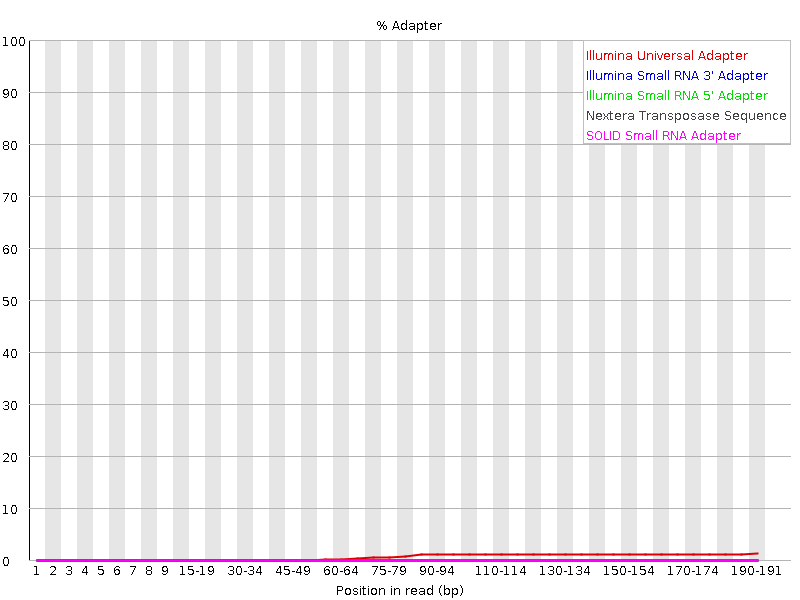
This graph shows us that this sequencing file has a low percentage (~2-3%) of adapter sequences in the reads, which means we do not need to trim any adapter sequences either.
When sequencing is poor(er) Quality
While the sequencing in this example is high quality this will not always be the case.
Here is an example of a good quality FastQC output and a bad quality FastQC output. The programe cutadapt can be used to filter poor quality reads and trim poor quality bases. See Genomics - Trimming and Filtering to learn more about trimming and filtering poor quality reads.
Nanopore quality control
Next we will assess the quality of the Nanopore raw reads. These are found in the file located at ~/cs_course/data/nano_fastq/ERR3152367_sub5.fastq.
Let us again view the first complete read in one of the files from our dataset by using head to look at the first four lines.
Move to the folder containing the Nanopore data:
cd ~/cs_course/data/nano_fastq/
Use head to look at the first four line of the fastq file:
head -n 4 ERR3152367_sub5.fastq
@ERR3152367.34573250 d8c83b24-b46e-4f1a-836f-768f835acf68 length=320
GGTTGGTTATGTGCATGTTTTCAGTTACATATTGCATCTGTGGGAGCATATTCTTGTTTATGGGTTATGTGTTGGTGGTTGCATGTGGTGTGTTGTTGTGTTAACAAGTGTGGAACCTGTTCATTGGGTTATGAACAACGACACAAGTGTTGCGTGTTGAGCTAGTTAACGTGTGTGTTGTTATTCTTCTGAACCAGTTAACTTATTTGTTTTGTTGGGTGTGAAGCAGTGGGCGTGAAGGTGAGCGATGAAGCGGCGTTGTTCTGTTGCGTGTTTGATTGTGTTGTGTTGCGTGAAGAAGCGTCGTTGTTGGGTGGTTC
+
$$##$$###$#%###%##$%%$$###$#$$#$%##%&$$$$$$%#$$$$#$%#%$##$#$%#%$$#$$$%#$$#$%$$$$$#$%#$#$%$$$##$%%#&$#$#$$$$$%$$%$$%%$$#"$#$$$#&$$$$$#$$$$$######$#$#$$###$%###$$$$%$$&%$$$#$#$$%#%$##$##%#$&$$$$$#$$$%$$$##%#%$##$%%$$#$$$$%#%$###$$$####%$%%$$'$$%$$$$$%$#$$&$$%$#$##$%%$$%$$%%$%&'##$##%$#$$%$###$$$$$#$$$$#$&%##$$#$$%$$$%###
This read is 320 bp, longer than the Illumina reads we looked at earlier. The length of a raw read from Nanopore sequencing varies depends on the length of the length of the DNA strand being sequenced.
Line 4 shows us the quality score of this read.
$$##$$###$#%###%##$%%$$###$#$$#$%##%&$$$$$$%#$$$$#$%#%$##$#$%#%$$#$$$%#$$#$%$$$$$#$%#$#$%$$$##$%%#&$#$#$$$$$%$$%$$%%$$#"$#$$$#&$$$$$#$$$$$######$#$#$$###$%###$$$$%$$&%$$$#$#$$%#%$##$##%#$&$$$$$#$$$%$$$##%#%$##$%%$$#$$$$%#%$###$$$####%$%%$$'$$%$$$$$%$#$$&$$%$#$##$%%$$%$$%%$%&'##$##%$#$$%$###$$$$$#$$$$#$&%##$$#$$%$$$%###
Based on the PHRED quality scores (see above for a reminder) we can see that the quality score of the bases in this read are between 1-10, which is lower than the Illumina sequencing above.
Instead of using FastQC we will use a program called NanoPlot, which is installed on the instance, to create some plots for the whole sequencing file. NanoPlot is specially built for Nanopore sequences.
Other programs for Nanopore QC
Another popular program for QC of Nanopore reads is PycoQC.
It produces similar plots to NanoPlot but will also give you information about the overall Nanopore sequencing run. In order to generate these, PycoQC uses a
sequencing summaryfile generated by the Nanopore sequencer (e.g. MiniION or PromethION).We are using NanoPlot because the
sequencing summarythat PycoQC needs is not avaiable for this dataset. You can see example output files from PycoQC here: Guppy-2.1.3_basecall-1D_DNA_barcode.html.
We first need to navigate to the qc directory we made earlier cs_course/analysis/qc.
cd ~/cs_course/analysis/qc/
We are now going to run NanoPlot with the raw Nanopore sequencing file.
First we can look at the help documenation for NanoPlot to see what options are available.
NanoPlot --help
NanoPlot Help Documentation
usage: NanoPlot [-h] [-v] [-t THREADS] [--verbose] [--store] [--raw] [--huge]m --downsample 10000 [-o OUTDIR] [-p PREFIX] [--tsv_stats] [--maxlength N] [--minlength N] [--drop_outliers] [--downsample N] [--loglength] [--percentqual] [--alength] [--minqual N] [--runtime_until N] [--readtype {1D,2D,1D2}] [--barcoded] [--no_supplementary] [-c COLOR] [-cm COLORMAP] [-f {eps,jpeg,jpg,pdf,pgf,png,ps,raw,rgba,svg,svgz,tif,tiff}] [--plots [{kde,hex,dot,pauvre} [{kde,hex,dot,pauvre} ...]]] [--listcolors] [--listcolormaps] [--no-N50] [--N50] [--title TITLE] [--font_scale FONT_SCALE] [--dpi DPI] [--hide_stats] (--fastq file [file ...] | --fasta file [file ...] | --fastq_rich file [file ...] | --fastq_minimal file [file ...] | --summary file [file ...] | --bam file [file ...] | --ubam file [file ...] | --cram file [file ...] | --pickle pickle | --feather file [file ...]) CREATES VARIOUS PLOTS FOR LONG READ SEQUENCING DATA. General options: -h, --help show the help and exit -v, --version Print version and exit. -t, --threads THREADS Set the allowed number of threads to be used by the script --verbose Write log messages also to terminal. --store Store the extracted data in a pickle file for future plotting. --raw Store the extracted data in tab separated file. --huge Input data is one very large file. -o, --outdir OUTDIR Specify directory in which output has to be created. -p, --prefix PREFIX Specify an optional prefix to be used for the output files. --tsv_stats Output the stats file as a properly formatted TSV. Options for filtering or transforming input prior to plotting: --maxlength N Hide reads longer than length specified. --minlength N Hide reads shorter than length specified. --drop_outliers Drop outlier reads with extreme long length. --downsample N Reduce dataset to N reads by random sampling. --loglength Additionally show logarithmic scaling of lengths in plots. --percentqual Use qualities as theoretical percent identities. --alength Use aligned read lengths rather than sequenced length (bam mode) --minqual N Drop reads with an average quality lower than specified. --runtime_until N Only take the N first hours of a run --readtype {1D,2D,1D2} Which read type to extract information about from summary. Options are 1D, 2D, 1D2 --barcoded Use if you want to split the summary file by barcode --no_supplementary Use if you want to remove supplementary alignments Options for customizing the plots created: -c, --color COLOR Specify a valid matplotlib color for the plots -cm, --colormap COLORMAP Specify a valid matplotlib colormap for the heatmap -f, --format {eps,jpeg,jpg,pdf,pgf,png,ps,raw,rgba,svg,svgz,tif,tiff} Specify the output format of the plots. --plots [{kde,hex,dot,pauvre} [{kde,hex,dot,pauvre} ...]] Specify which bivariate plots have to be made. --listcolors List the colors which are available for plotting and exit. --listcolormaps List the colors which are available for plotting and exit. --no-N50 Hide the N50 mark in the read length histogram --N50 Show the N50 mark in the read length histogram --title TITLE Add a title to all plots, requires quoting if using spaces --font_scale FONT_SCALE Scale the font of the plots by a factor --dpi DPI Set the dpi for saving images --hide_stats Not adding Pearson R stats in some bivariate plots Input data sources, one of these is required.: --fastq file [file ...] Data is in one or more default fastq file(s). --fasta file [file ...] Data is in one or more fasta file(s). --fastq_rich file [file ...] Data is in one or more fastq file(s) generated by albacore, MinKNOW or guppy with additional information concerning channel and time. --fastq_minimal file [file ...] Data is in one or more fastq file(s) generated by albacore, MinKNOW or guppy with additional information concerning channel and time. Is extracted swiftly without elaborate checks. --summary file [file ...] Data is in one or more summary file(s) generated by albacore or guppy. --bam file [file ...] Data is in one or more sorted bam file(s). --ubam file [file ...] Data is in one or more unmapped bam file(s). --cram file [file ...] Data is in one or more sorted cram file(s). --pickle pickle Data is a pickle file stored earlier. --feather file [file ...] Data is in one or more feather file(s). EXAMPLES: NanoPlot --summary sequencing_summary.txt --loglength -o summary-plots-log-transformed NanoPlot -t 2 --fastq reads1.fastq.gz reads2.fastq.gz --maxlength 40000 --plots hex dot NanoPlot --color yellow --bam alignment1.bam alignment2.bam alignment3.bam --downsample 10000
We will use four flags when we run the NanoPlot command:
We also use --outdir to specify an output directory. We’re also going to use the flag --loglength to produce plots with a log scale and finally we’re going to use --threads to run the program on more than one thread to speed it up.
-
--fastqto specify the filetype and file to analyse. The raw Nanopore data is in the location/cs_workshop/data/nano_fastq/ERR3152367_sub5.fastqand we will use this full absolute path in the NanoPlot command. -
--outdirto specify the where the output files should be written. We are going to specifynano_qcso that NanoPlot will create a new directory in our current directory (qc) and write its output files to it. Note: with NanoPlot you don’t need to create this directory before running the command. -
--threadsspecifies how many threads to run the program on (more threads = more compute power = faster). We will specify 4 to indicate that four threads should be used. -
--loglengthspecifies that we want plots with a log scale.
NanoPlot --fastq ~/cs_course/data/nano_fastq/ERR3152367_sub5.fastq --outdir nano_qc --threads 4 --loglength
Now we have the command set up we can press enter and wait for NanoPlot to finish.
This will take a couple of minutes. You will know it is finished once your cursor has returned (i.e. you can type in the terminal again).
Once NanoPlot has finished we can have a look at the output.
First we need to navigate into the nano_qc directory NanoPlot created, then list the files.
cd nano_qc
ls
LengthvsQualityScatterPlot_dot.html LengthvsQualityScatterPlot_loglength_kde.png Non_weightedLogTransformed_HistogramReadlength.png
LengthvsQualityScatterPlot_dot.png NanoPlot_20221005_1630.log WeightedHistogramReadlength.html
LengthvsQualityScatterPlot_kde.html NanoPlot-report.html WeightedHistogramReadlength.png
LengthvsQualityScatterPlot_kde.png NanoStats.txt WeightedLogTransformed_HistogramReadlength.html
LengthvsQualityScatterPlot_loglength_dot.html Non_weightedHistogramReadlength.html WeightedLogTransformed_HistogramReadlength.png
LengthvsQualityScatterPlot_loglength_dot.png Non_weightedHistogramReadlength.png Yield_By_Length.html
LengthvsQualityScatterPlot_loglength_kde.html Non_weightedLogTransformed_HistogramReadlength.html Yield_By_Length.png
We can see that NanoPlot has generated a lot of different files.
Like before, we can’t view most of these files in our terminal as we can’t open images or HTML files. Instead we’ll download the core information to our own computer.
Luckily, the NanoPlot-report.html file contains all of the plots and information held in the other files so we only need to download that one onto our local computer using scp.
Use a terminal that is not logged into the cloud instance and ensure you are in your cloudspan directory. You may have one from earlier. If you do not, use the instructions above to start one.
Use scp to copy the file - the command will look something like:
scp -i login-key-instanceNNN.pem csuser@instanceNNN.cloud-span.aws.york.ac.uk:~/cs_course/analysis/qc/nano_qc/NanoPlot-report.html .
Remember to replace NNN with the instance number specific to you. As the file is downloading you will see an output like:
scp -i login-key-instanceNNN.pem csuser@instanceNNN.cloud-span.aws.york.ac.uk:~/cs_course/analysis/qc/nano_qc/NanoPlot-report.html .
NanoPlot-report.html 100% 3281KB 2.3MB/s 00:01
Once the file has downloaded File Explorer (Windows) or Finder (Mac) to find the file and open it - it should open up in your browser.
Help!
If you had trouble downloading and viewing the file you can view it here: NanoPlot-report.html
In the report we can view summary statistics followed by plots showing the distribution of read lengths and the read length vs average read quality.
Looking at the summary statistics table answer the following questions:
Exercise 1:
- How many sequences are in this file?
- How many bases are there in this entire file?
- What is the length of the longest read in the file and its associated mean quality score?
Solution
- There are 692,758 sequences (also known as reads) in this file
- There are 3,082,258,211 bases (bp) in total in this FASTQ file
- The longest read in this file is 413,847 bp and it has a mean quality score of 3.7
Quality Encodings Vary
Note that not all sequencing machines use the same encoding for quality. So
#might not always mean 3, a poor quality score.This means it’s essential that you know which sequencing platform was used to generate your data, so that you can tell your quality control program which encoding to use. If you choose the wrong encoding, you run the risk of throwing away good reads or (even worse) not throwing away bad reads! Nanopore quality encodings are no exception. You can read more about the differences with Nanopore sequencing here: EPI2ME - Quality Scores.
N50
The N50 length is a useful statistic when looking at sequences of varying length as it indicates that 50% of the total sequence is in reads (i.e. chunks) that are that size or larger.
For this FASTQ file 50% of the total bases are in reads that have a length of 5,373 bp or longer.
See the webpage What’s N50? for a good explanation. We will be coming back to this statistic in more detail when we get to the assembly step.
We can also look at some of the plots produced by NanoPlot.
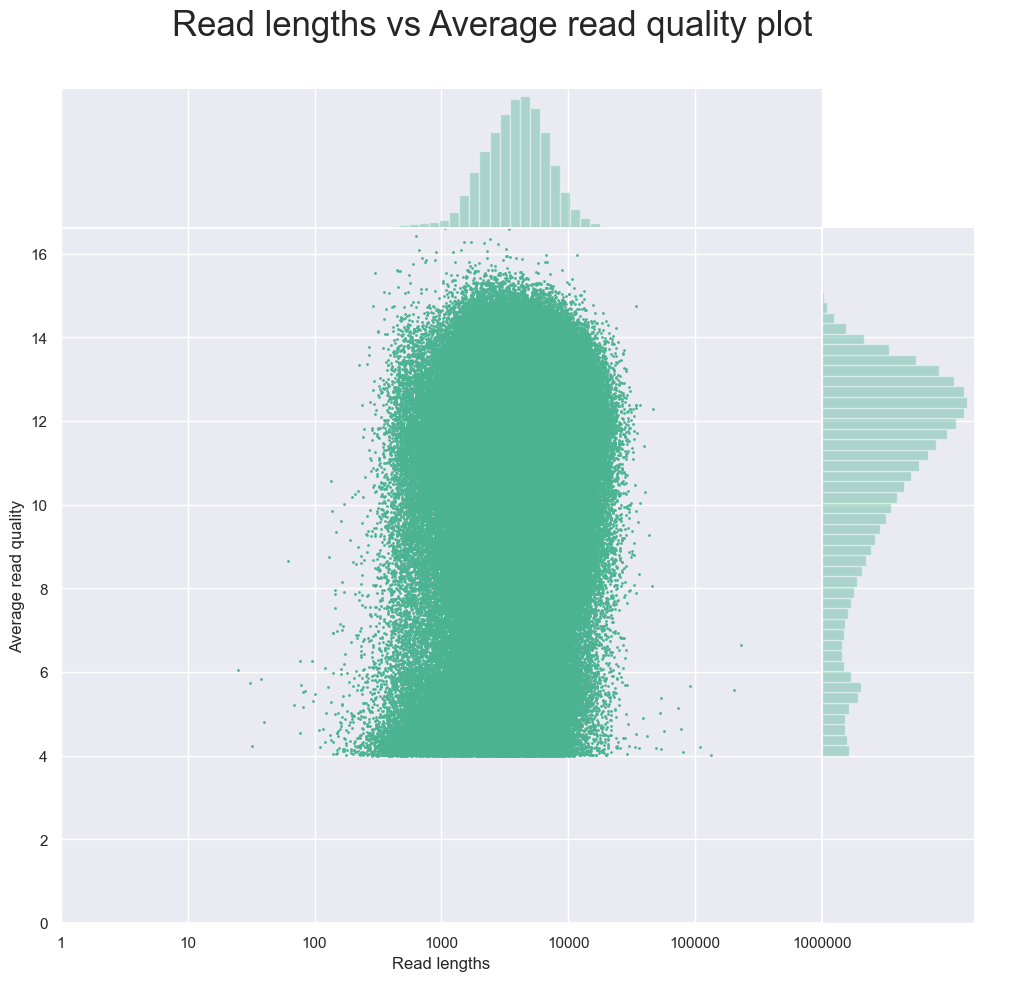
One useful plot is the plot titled “Read lengths vs Average read quality plot using dots after log transformation of read lengths”.
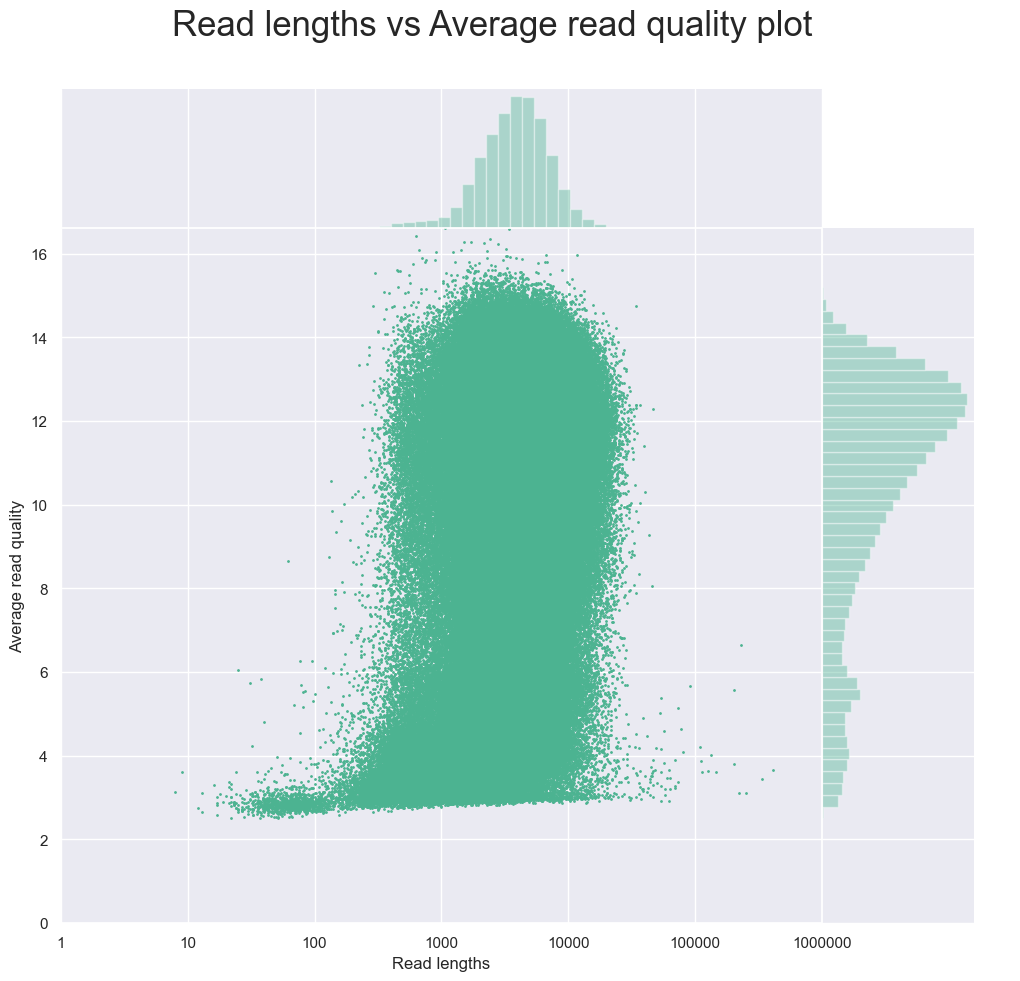
This plot shows the average quality of the sequence against the read lengths. We can see that the majority of the sequences have a quality score at least 4, and low quality scores come from very short reads. This means that for this dataset we should remove those with a lower quality score in order to improve the overall quality of the raw sequences before assembling the metagenome.
Filtering Nanopore sequences by quality
We can use the program Seqkit (which contains many tools for FASTQ/A file manipulation) to filter our reads. We will be using the command seqkit seq to create a new file containing only the sequences with an average quality above a certain value.
After returning to our home directory, we can view the seqkit seq help documentation with the following command:
cd ~/cs_course/
seqkit seq -h
Seqkit seq help documentation
transform sequences (extract ID, filter by length, remove gaps...) Usage: seqkit seq [flags] Flags: -k, --color colorize sequences - to be piped into "less -R" -p, --complement complement sequence, flag '-v' is recommended to switch on --dna2rna DNA to RNA -G, --gap-letters string gap letters (default "- \t.") -h, --help help for seq -l, --lower-case print sequences in lower case -M, --max-len int only print sequences shorter than the maximum length (-1 for no limit) (default -1) -R, --max-qual float only print sequences with average quality less than this limit (-1 for no limit) (default -1) -m, --min-len int only print sequences longer than the minimum length (-1 for no limit) (default -1) -Q, --min-qual float only print sequences with average quality qreater or equal than this limit (-1 for no limit) (default -1) -n, --name only print names -i, --only-id print ID instead of full head -q, --qual only print qualities -b, --qual-ascii-base int ASCII BASE, 33 for Phred+33 (default 33) -g, --remove-gaps remove gaps -r, --reverse reverse sequence --rna2dna RNA to DNA -s, --seq only print sequences -u, --upper-case print sequences in upper case -v, --validate-seq validate bases according to the alphabet -V, --validate-seq-length int length of sequence to validate (0 for whole seq) (default 10000) Global Flags: --alphabet-guess-seq-length int length of sequence prefix of the first FASTA record based on which seqkit guesses the sequence type (0 for whole seq) (default 10000) --id-ncbi FASTA head is NCBI-style, e.g. >gi|110645304|ref|NC_002516.2| Pseud... --id-regexp string regular expression for parsing ID (default "^(\\S+)\\s?") --infile-list string file of input files list (one file per line), if given, they are appended to files from cli arguments -w, --line-width int line width when outputting FASTA format (0 for no wrap) (default 60) -o, --out-file string out file ("-" for stdout, suffix .gz for gzipped out) (default "-") --quiet be quiet and do not show extra information -t, --seq-type string sequence type (dna|rna|protein|unlimit|auto) (for auto, it automatically detect by the first sequence) (default "auto") -j, --threads int number of CPUs. can also set with environment variable SEQKIT_THREADS) (default 4)
From this we can see that the flag -Q will “only print sequences with average quality qreater or equal than this limit (-1 for no limit) (default -1)”.
From the plot above we identified that many of the lower quality reads below 4 were shorter more here so we should set the minimum limit to 4.
seqkit seq -Q 4 data/nano_fastq/ERR3152367_sub5.fastq > data/nano_fastq/ERR3152367_sub5_filtered.fastq
In the command above we use redirection (>) to generate a new file data/nano_fastq/ERR3152367_sub5_filtered.fastq containing only the reads with an average quality of 4 or above.
We can now re-run NanoPlot on the filtered file to see how it has changed.
cd analysis/qc/
NanoPlot --fastq ~/cs_course/data/nano_fastq/ERR3152367_sub5_filtered.fastq --outdir nano_qc_filt --threads 4 --loglength
Once again, wait for the command to finish and then scp the NanoPlot-report.html to your local computer.
Help!
If you had trouble downloading the file you can view it here: NanoPlot-filtered-report.html
Compare the NanoPlot statistics of the Nanopore raw reads before filtering and after filtering and answer the questions below.
Exercise 2:
- How many reads have been removed by filtering?
- How many bases have been removed by filtering?
- What is the length of the new longest read and its associated average quality score?
Solution
- Initially there were 692,758 reads in the filtered file there are 666,597 reads so 26,161 reads have been removed by the quality filtering
- Initially there were 3,082,258,211 bases and after filtering there are 3,023,658,929 base which means filtering has removed 58,599,282 bases
- The longest read in the filtered file is 229,804bp and it has a mean quality score of 6.7
Key Points
Quality encodings vary across sequencing platforms.
It is important to know the quality of our data to be able to make decisions in the subsequent steps.
Data cleaning is essential at the beginning of metagenomics workflows.
Due to differences in the sequencing technology Nanopore data must be handled differently.