Redirection
Overview
Teaching: 30 min
Exercises: 15 minQuestions
How can I search within files?
How can I combine existing commands to do new things?
Objectives
Employ the
grepcommand to search for information within files.Print the results of a command to a file.
Construct command pipelines with two or more stages.
Use
forloops to run the same command for several input files.
Searching files
We discussed in a previous episode how to search within a file using less. We can also
search within files without even opening them, using grep. grep is a command-line
utility for searching plain-text files for lines matching a specific set of
characters (sometimes called a string) or a particular pattern
(which can be specified using something called regular expressions). We’re not going to work with
regular expressions in this lesson, and are instead going to specify the strings
we are searching for.
Let’s give it a try!
Nucleotide abbreviations
The four nucleotides that appear in DNA are abbreviated
A,C,TandG. Unknown nucleotides are represented with the letterN. AnNappearing in a sequencing file represents a position where the sequencing machine was not able to confidently determine the nucleotide in that position. You can think of anNas being aNy nucleotide at that position in the DNA sequence.
We’ll search for strings inside of our fastq files. Let’s first make sure we are in the correct directory:
$ cd ~/shell_data/untrimmed_fastq
Suppose we want to see how many reads in our file have really bad segments containing 10 consecutive unknown nucleotides (Ns).
Determining quality
In this lesson, we’re going to be manually searching for strings of
Ns within our sequence results to illustrate some principles of file searching. It can be really useful to do this type of searching to get a feel for the quality of your sequencing results, however, in your research you will most likely use a bioinformatics tool that has a built-in program for filtering out low-quality reads. You’ll learn how to use one such tool in a Genomics lesson.
Let’s search for the string NNNNNNNNNN in the SRR098026 file:
$ grep NNNNNNNNNN SRR098026.fastq
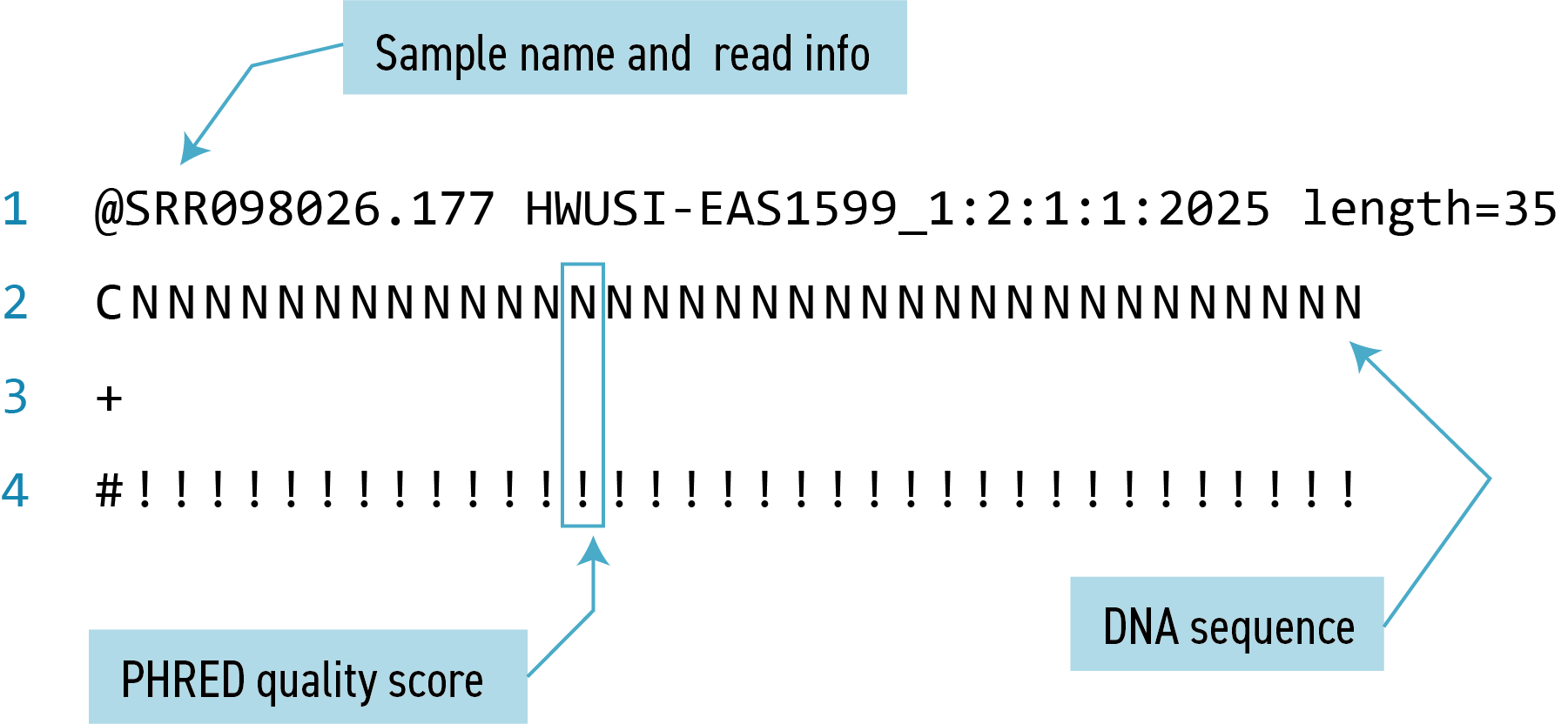
This command returns a lot of output to the terminal. Every single line in the SRR098026 file that contains at least 10 consecutive Ns is printed to the terminal, regardless of how long or short the file is. We may be interested not only in the actual sequence which contains this string, but in the name (or identifier) of that sequence. Think back to the FASTQ format we discussed previously - if you need a reminder, you can click to reveal one below.
Hint
To get all of the information about this read, we will return the line immediately before each match and the two lines immediately after each match.
We can use the -B argument for grep to return a specific number of lines before
each match. The -A argument returns a specific number of lines after each matching line. Here we want the line before and the two lines after each
matching line, so we add -B1 -A2 to our grep command:
$ grep -B1 -A2 NNNNNNNNNN SRR098026.fastq
One of the sets of lines returned by this command is:
@SRR098026.177 HWUSI-EAS1599_1:2:1:1:2025 length=35
CNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
+SRR098026.177 HWUSI-EAS1599_1:2:1:1:2025 length=35
#!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!
Exercise
Search for the sequence
GNATNACCACTTCCin theSRR098026.fastqfile. Have your search return all matching lines and the name (or identifier) for each sequence that contains a match. What is the output?Search for the sequence
AAGTTin both FASTQ files. Have your search return all matching lines and the name (or identifier) for each sequence that contains a match. How many matching sequences do you get?Share your answers on the forum.
Solution
grep -B1 GNATNACCACTTCC SRR098026.fastq@SRR098026.245 HWUSI-EAS1599_1:2:1:2:801 length=35 GNATNACCACTTCCAGTGCTGANNNNNNNGGGATG
grep -B1 AAGTT *.fastqSRR097977.fastq-@SRR097977.11 209DTAAXX_Lenski2_1_7:8:3:247:351 length=36 SRR097977.fastq:GATTGCTTTAATGAAAAAGTCATATAAGTTGCCATG -- SRR097977.fastq-@SRR097977.67 209DTAAXX_Lenski2_1_7:8:3:544:566 length=36 SRR097977.fastq:TTGTCCACGCTTTTCTATGTAAAGTTTATTTGCTTT -- SRR097977.fastq-@SRR097977.68 209DTAAXX_Lenski2_1_7:8:3:724:110 length=36 SRR097977.fastq:TGAAGCCTGCTTTTTTATACTAAGTTTGCATTATAA -- SRR097977.fastq-@SRR097977.80 209DTAAXX_Lenski2_1_7:8:3:258:281 length=36 SRR097977.fastq:GTGGCGCTGCTGCATAAGTTGGGTTATCAGGTCGTT -- SRR097977.fastq-@SRR097977.92 209DTAAXX_Lenski2_1_7:8:3:353:318 length=36 SRR097977.fastq:GGCAAAATGGTCCTCCAGCCAGGCCAGAAGCAAGTT -- SRR097977.fastq-@SRR097977.139 209DTAAXX_Lenski2_1_7:8:3:703:655 length=36 SRR097977.fastq:TTTATTTGTAAAGTTTTGTTGAAATAAGGGTTGTAA -- SRR097977.fastq-@SRR097977.238 209DTAAXX_Lenski2_1_7:8:3:592:919 length=36 SRR097977.fastq:TTCTTACCATCCTGAAGTTTTTTCATCTTCCCTGAT -- SRR098026.fastq-@SRR098026.158 HWUSI-EAS1599_1:2:1:1:1505 length=35 SRR098026.fastq:GNNNNNNNNCAAAGTTGATCNNNNNNNNNTGTGCG
Redirecting output
grep allowed us to identify sequences in our FASTQ files that match a particular pattern.
All of these sequences were printed to our terminal screen, but in order to work with these
sequences and perform other operations on them, we will need to capture that output in some
way.
We can do this with something called “redirection”. The idea is that we are taking what would ordinarily be printed to the terminal screen and redirecting it to another location. In our case, we want to print this information to a file so that we can look at it later and use other commands to analyse this data.
The command for redirecting output to a file is >.
Let’s try out this command and copy all the records (including all four lines of each record)
in our FASTQ files that contain
‘NNNNNNNNNN’ to another file called bad_reads.txt.
$ grep -B1 -A2 NNNNNNNNNN SRR098026.fastq > bad_reads.txt
File extensions
You might be confused about why we’re naming our output file with a
.txtextension. After all, it will be holding FASTQ formatted data that we’re extracting from our FASTQ files. Won’t it also be a FASTQ file? The answer is, yes - it will be a FASTQ file and it would make sense to name it with a.fastqextension. However, using a.fastqextension will lead us to problems when we move to using wildcards later in this episode. We’ll point out where this becomes important. For now, it’s good that you’re thinking about file extensions!
The prompt should sit there a little bit, and then it should look like nothing
happened. But type ls. You should see a new file called bad_reads.txt.
We can check the number of lines in our new file using a command called wc.
wc stands for word count. This command counts the number of words, lines, and characters
in a file. The FASTQ file may change over time, so given the potential for updates,
make sure your file matches your instructor’s output.
As of Nov. 2021, wc gives the following output:
$ wc bad_reads.txt
537 1073 23217 bad_reads.txt
This will tell us the number of lines, words and characters in the file. If we
want only the number of lines, we can use the -l flag for lines.
$ wc -l bad_reads.txt
537 bad_reads.txt
Exercise
Try these questions on your own first. Then, if you are attending an instructor-led workshop, discuss your answers in your breakout room. Nominate someone from your group to feed back on the forum.
- How many sequences are there in
SRR098026.fastq? Remember that every sequence is formed by four lines.- How many sequences in
SRR098026.fastqcontain at least 3 consecutive Ns?Solution
1.
$ wc -l SRR098026.fastq996Now you can divide this number by four to get the number of sequences in your fastq file
2.
$ grep NNN SRR098026.fastq > bad_reads.txt $ wc -l bad_reads.txt249
We might want to search multiple FASTQ files for sequences that match our search pattern.
However, we need to be careful, because each time we use the > command to redirect output
to a file, the new output will replace the output that was already present in the file.
This is called “overwriting” and, just like you don’t want to overwrite your video recording
of your kid’s first birthday party, you also want to avoid overwriting your data files.
$ grep -B1 -A2 NNNNNNNNNN SRR098026.fastq > bad_reads.txt
$ wc -l bad_reads.txt
537 bad_reads.txt
$ grep -B1 -A2 NNNNNNNNNN SRR097977.fastq > bad_reads.txt
$ wc -l bad_reads.txt
0 bad_reads.txt
Here, the output of our second call to wc shows that we no longer have any lines in our bad_reads.txt file. This is
because the second file we searched (SRR097977.fastq) does not contain any lines that match our
search sequence. So our file was overwritten and is now empty.
We can avoid overwriting our files by using the command >>. >> is known as the “append redirect” and will
append new output to the end of a file, rather than overwriting it.
$ grep -B1 -A2 NNNNNNNNNN SRR098026.fastq > bad_reads.txt
$ wc -l bad_reads.txt
537 bad_reads.txt
$ grep -B1 -A2 NNNNNNNNNN SRR097977.fastq >> bad_reads.txt
$ wc -l bad_reads.txt
537 bad_reads.txt
The output of our second call to wc shows that we have not overwritten our original data.
We can also do this with a single line of code by using a wildcard:
$ grep -B1 -A2 NNNNNNNNNN *.fastq > bad_reads.txt
$ wc -l bad_reads.txt
537 bad_reads.txt
Since we might have multiple different criteria we want to search for,
creating a new output file each time has the potential to clutter up our workspace. We also
thus far haven’t been interested in the actual contents of those files, only in the number of
reads that we’ve found. We created the files to store the reads and then counted the lines in
the file to see how many reads matched our criteria. There’s a way to do this, however, that
doesn’t require us to create these intermediate files - the pipe command (|).
This is probably not a key on
your keyboard you use very much, so let’s all take a minute to find that key. For the standard QWERTY keyboard
layout, the | character can be found using the key combination
- Shift+\
What | does is take the output that is scrolling by on the terminal and uses that output as input to another command.
When our output was scrolling by, we might have wished we could slow it down and
look at it, like we can with less. Well it turns out that we can! We can redirect our output
from our grep call through the less command.
$ grep -B1 -A2 NNNNNNNNNN SRR098026.fastq | less
We can now see the output from our grep call within the less interface. We can use the up and down arrows
to scroll through the output and use q to exit less.
If we don’t want to create a file before counting lines of output from our grep search, we could directly pipe
the output of the grep search to the command wc -l. This can be helpful for investigating your output if you are not sure
you would like to save it to a file.
$ grep -B1 -A2 NNNNNNNNNN SRR098026.fastq | wc -l
Custom
grepcontrolUse
man grepto read more about other options to customize the output ofgrepincluding extended options, anchoring characters, and much more.
Redirecting output is often not intuitive, and can take some time to get used to. Once you’re comfortable with redirection, however, you’ll be able to combine any number of commands to do all sorts of exciting things with your data!
None of the command line programs we’ve been learning do anything all that impressive on their own, but when you start chaining them together, you can do some really powerful things very efficiently.
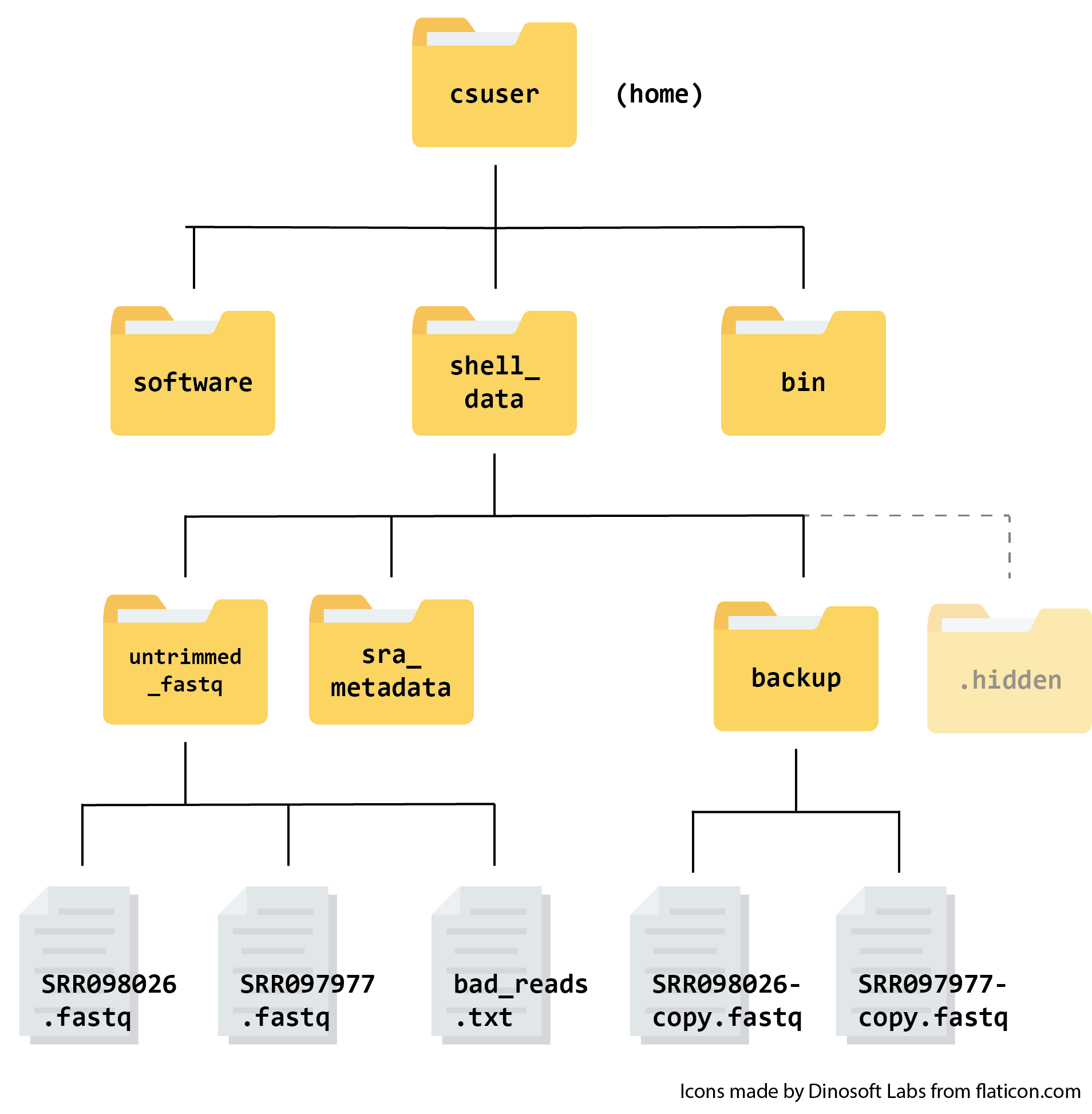
Here is what your file structure should look like at the end of this episode:
Key Points
grepis a powerful search tool with many options for customization.
>,>>, and|are different ways of redirecting output.
command > fileredirects a command’s output to a file.
command >> fileredirects a command’s output to a file without overwriting the existing contents of the file.
command_1 | command_2redirects the output of the first command as input to the second command.