The paper
This course uses data from a 2022 paper published in BMC Environmental Microbiome titled In-depth characterization of denitrifier communities across different soil ecosystems in the tundra.
The paper characterises microbial communities across 43 mountain tundra sites in northern Finland with focus on their role in nitrous oxide cycling. Each site was classified as being barren, heathland, meadow or fen.
Environmental information such as elevation, soil moisture and N2O fluxes was gathered for each site. DNA from each site was extracted and sequenced using Illumina technology, giving “short reads”. Two sites were additionally sequenced using a Nanopore MinION, giving “long reads”. In this course we will focus on these two sites as they have both long and short read data available.
The sites
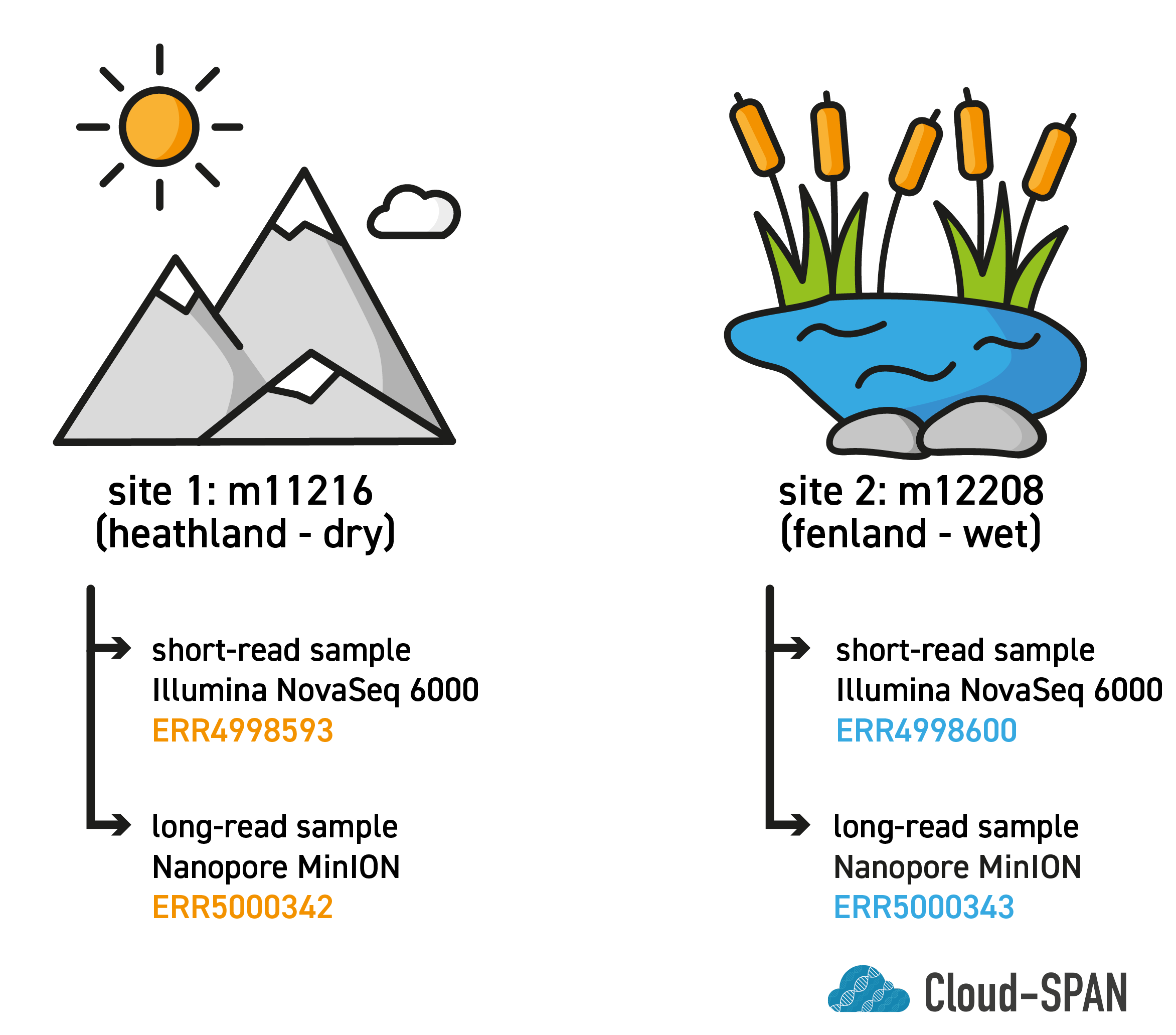
The main site we will be looking at is site m11216, a heathland site with elevation 812.7 metres above sea level. We have two samples for this site: a short-read sample called ERR4998593 and a long-read sample called ERR5000342. These are the samples we will be working with most and using in our metagenomics analysis workflow.
Towards the end of the course we will compare this site with site m12208, a fenland site with elevation 706.3m above sea level. We will not be using the samples associated with this site during the course but we will use the outputs of their analysis for comparison with our own.