Other Resources
Overview
Teaching: 15 min
Exercises: 0 minQuestions
Where can I find other metagenomic resources?
Objectives
Know some places where to find more information.
Other resources
We hope that you have found these lessons useful. However, this course is only the start! There is lots more you can learn about analysing and understanding metagenomics, and how to apply these techniques to your own dataset.
A quick roundup of useful learning resources:
- This Phyloseq tutorial contains many more examples of metagenome data manipulation.
- A’nvio developer Meren has a blog entry about History of metagenomics
- The same developer also has several videos explaining topics such as Metapangenomics: A nexus between pangenomes and metagenomes, The power of metagenomic read recruitment and Genome-resolved metagenomics: key concepts in reconstructing genomes from metagenomes.
There are also some great non-command line resources out there that may be great to get you started. Here are some of our suggestions, although this is by no means an exhaustive list.
MG-RAST
A useful and easy to use resource is MG-RAST, an online metagenomic platform where you can upload your raw data with its corresponding metadata and get a full taxonomic analysis of it. MG-RAST is also a big repository of available data for future experiments.
A downside of this resource is that it is not possible to modify the steps and parameters in the MG-RAST workflow, meaning there is less flexibility for implementing your preferred analysis tools.
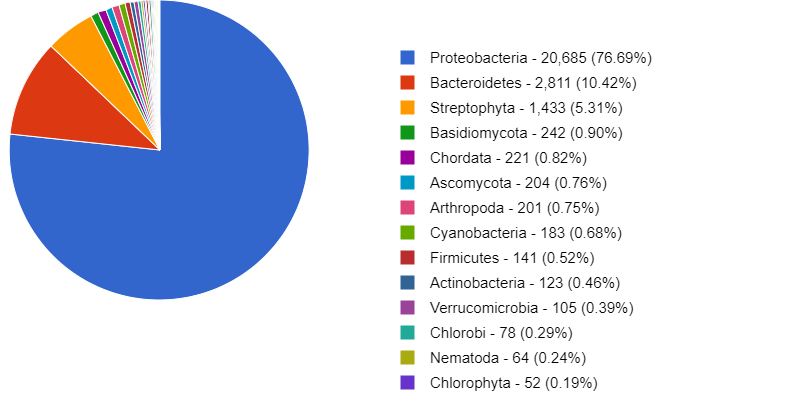
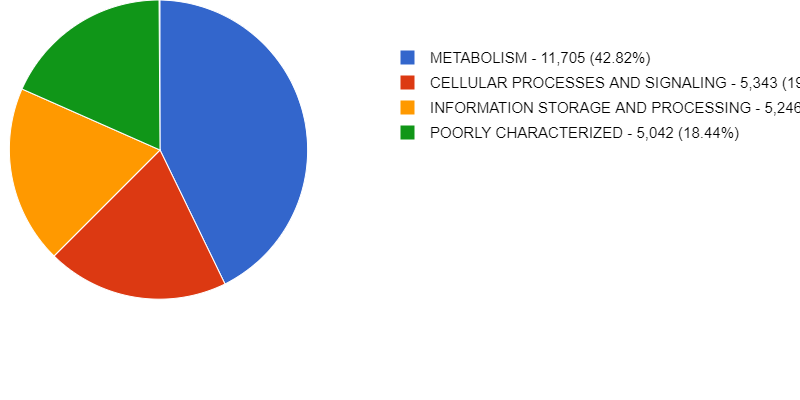
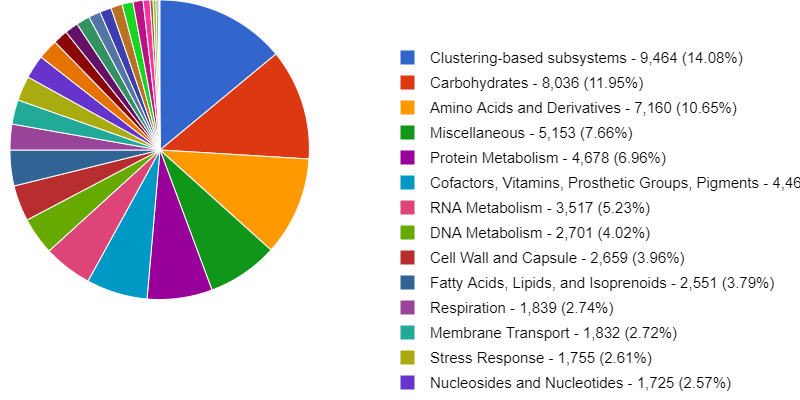
Here is an example of some of the figures generated by MG-RAST. It annotates the metabolic function and can give you an idea of what your organisms/ individual MAGs are capable of metabolising.
MG-RAST has is a very useful tool for taking a quick look at your data, as well as storing and sharing it.
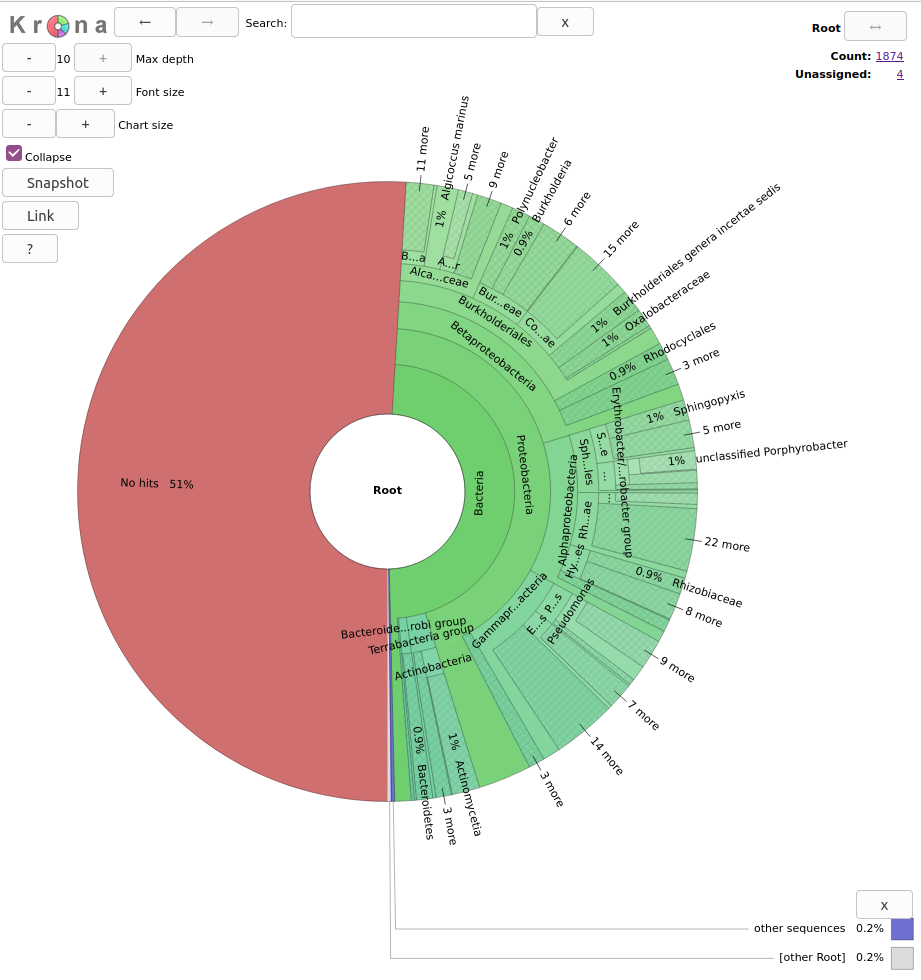
Krona for visualization
Krona is a hierarchical data visualisation software. It allows data to be explored with zooming, multi-layered pie charts and includes support for several bioinformatics tools and raw data formats. Using Krona you can generate HTML reports on the taxonomy in your metagenome like the one below.
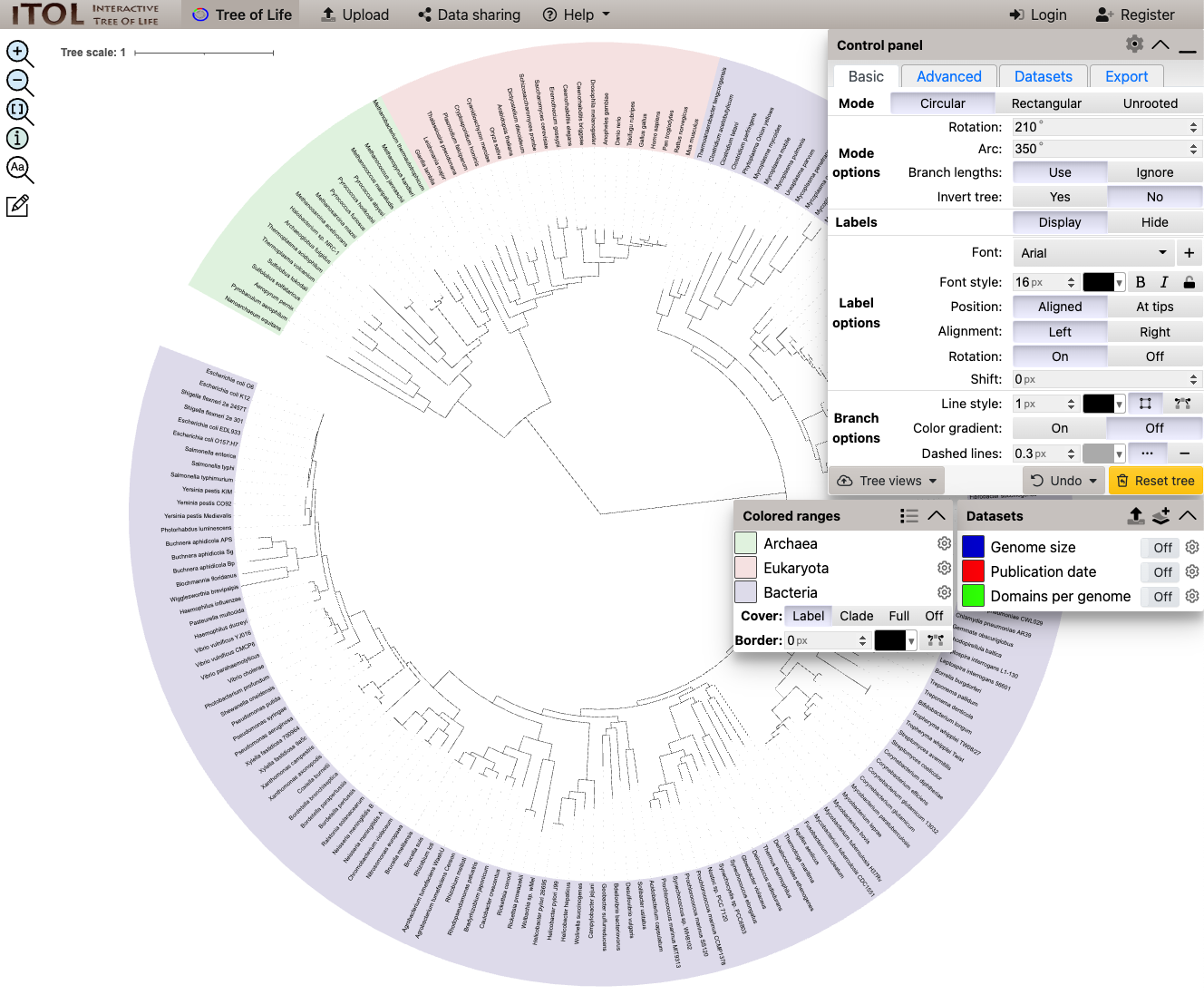
iTOL, the interactive tree of life
iTOL is a free website which can be used to generate publication quality phylogenetic trees. Below shows the interface you can use to customise parts of the plot. There are also several video tutorials to show you how to get started. You can also share your trees with other people easily.
This tool is useful if you want to make a tree based on the 16S sequence for several genomes in your metagenome (annotated by Prokka), or if you are interested in a particular set of genes within a genome.
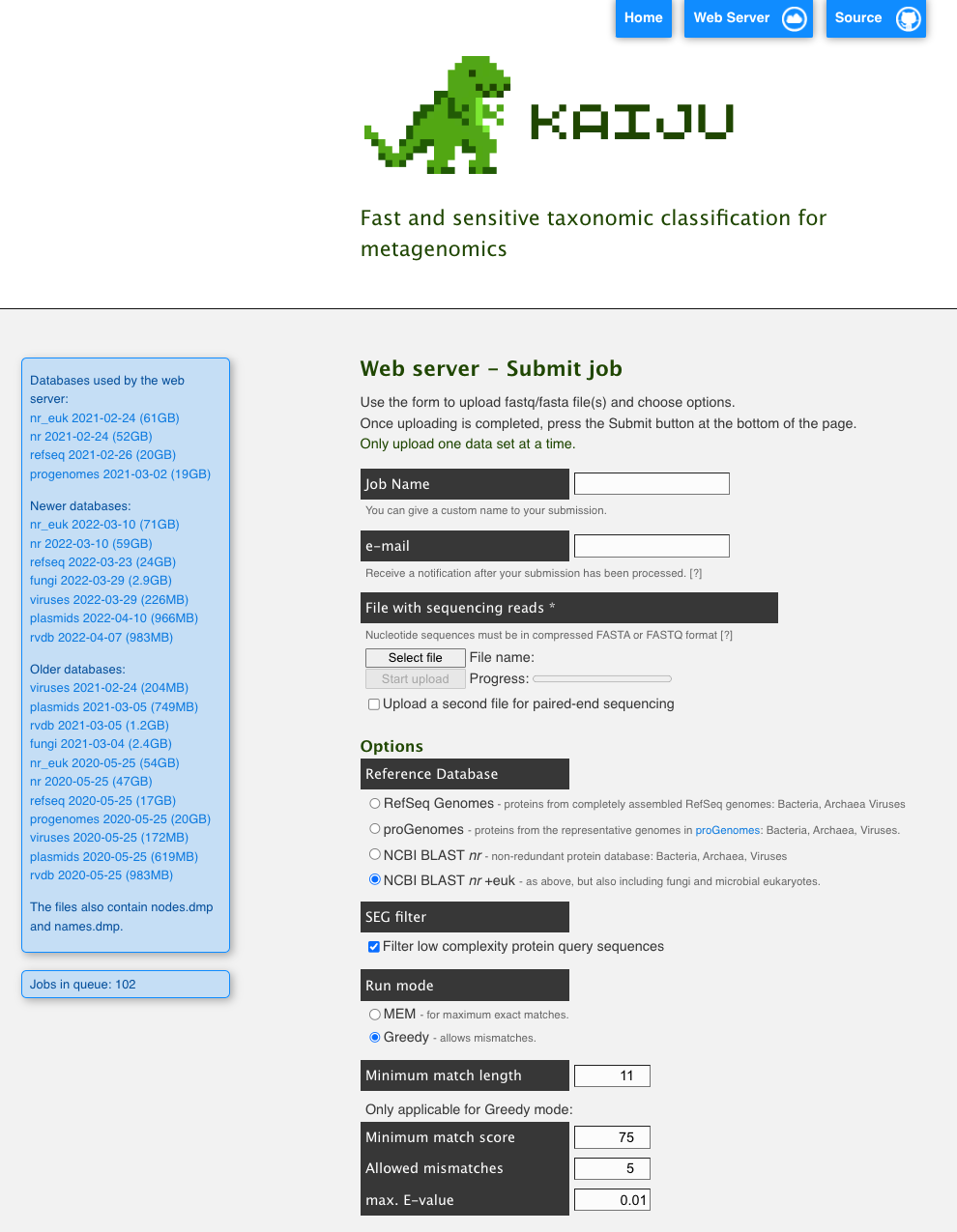
KAIJU - alternative classification tool
Kraken2 (the tool we used) is a very popular command line tool for annotating the short reads in your metagenome. KAIJU is another popular tool which has both a command line version and a web based version. It has databases for microbial eukaryotes and viruses as well as bacterial and archaeal refseq genomes.
Here’s what the web version looks like:
MetaPhlAn for taxonomic profiling
MetaPhlAn 4 is another very popular tool for profiling taxonomy in microbial communities. The latest associated paper is here.
One benefit of MetaPhlAn is that it has an actively updated github repository as well as an active support forum.It was developed as part of the BioBakery, which also has many other excellent tools and resources.
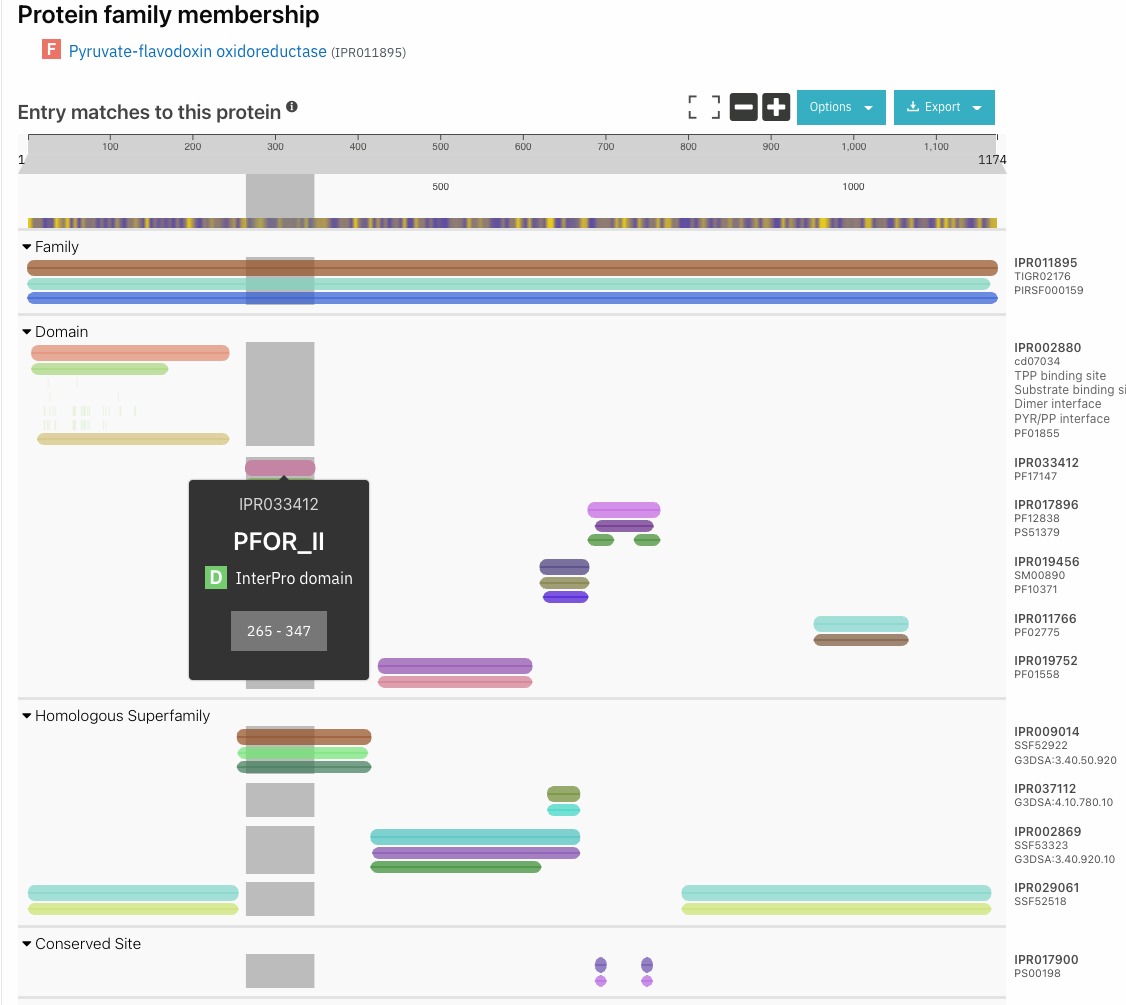
Identifying functional domains with InterProScan
If you are interested in identifying the functional domain of proteins, InterProScan should be the first tool you use. It has both a command line version for annotating many proteins at once, and a website version for predicting smaller numbers of proteins. It produces an exportable graphical output as shown below.
Recommended reading:
Review articles can be a useful source of information and some contain comprehensive lists of different tools and databases to use for your metagenomic analysis. Here are two we recommend:
Key Points
Metagenomes are complicated to analyse. Luckily, there are lots of tools available!
The best tools to use change frequently. Do your research when looking for the most suitable tools to use for your analysis.