This extra lesson is optional and requires you to download software to your own PC or laptop.
Optional Extra: Alignment in IGV
Installing IGV
For this lesson you will need to download IGV from the Broad Institute’s software page, double-click the
.zipfile to unzip it, and then drag the program into your Applications folder.
Viewing with IGV
IGV is a stand-alone browser, which has the advantage of being installed locally and providing fast access. Web-based genome browsers, like Ensembl or the UCSC browser, are slower, but provide more functionality. They not only allow for more polished and flexible visualization, but also provide easy access to a wealth of annotations and external data sources. This makes it straightforward to relate your data with information about repeat regions, known genes, epigenetic features or areas of cross-species conservation, to name just a few.
In order to use IGV, we will need to transfer some files to our local machine. We know how to do this with scp.
Open a new tab in your terminal window and create a new folder. We’ll put this folder on our Desktop for
demonstration purposes, but in general you should avoide proliferating folders and files on your Desktop and
instead organise files within a directory structure like we’ve been using in our cs_course directory.
$ mkdir ~/Desktop/files_for_igv
$ cd ~/Desktop/files_for_igv
Now we will transfer our files to that new directory. Remember to replace the text between the @ and the :
with your AWS instance number. The commands to scp always go in the terminal window that is connected to your
local computer (not your AWS instance).
$ scp csuser@ec2-34-203-203-131.compute-1.amazonaws.com:~/cs_course/SRR2584866.aligned.sorted.bam ~/Desktop/files_for_igv
$ scp csuser@ec2-34-203-203-131.compute-1.amazonaws.com:~/cs_course/SRR2584866.aligned.sorted.bam.bai ~/Desktop/files_for_igv
$ scp csuser@ec2-34-203-203-131.compute-1.amazonaws.com:~/cs_course/data/ecoli_rel606.fasta ~/Desktop/files_for_igv
$ scp csuser@ec2-34-203-203-131.compute-1.amazonaws.com:~/cs_course/SRR2584866_final_variants.vcf ~/Desktop/files_for_igv
You will need to type the password for your AWS instance each time you call scp.
Next, we need to open the IGV software. If you haven’t done so already, you can
- Open IGV.
- Load our reference genome file (
ecoli_rel606.fasta) into IGV using the “Load Genomes from File…“ option under the “Genomes” pull-down menu. - Load our BAM file (
SRR2584866.aligned.sorted.bam) using the “Load from File…“ option under the “File” pull-down menu. - Do the same with our VCF file (
SRR2584866_final_variants.vcf).
Your IGV browser should look like the screenshot below:
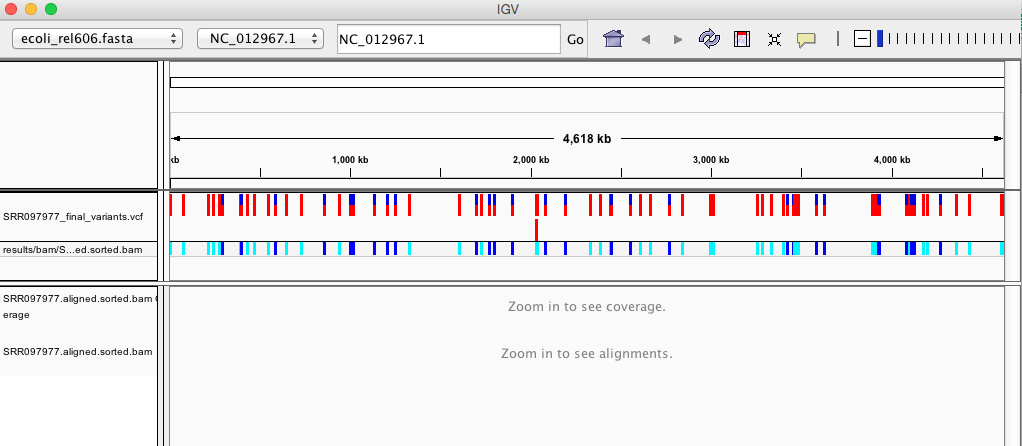
There should be two tracks: one coresponding to our BAM file and the other for our VCF file.
In the VCF track, each bar across the top of the plot shows the allele fraction for a single locus. The second bar shows the genotypes for each locus in each sample. We only have one sample called here, so we only see a single line. Dark blue = heterozygous, Cyan = homozygous variant, Grey = reference. Filtered entries are transparent.
Zoom in to inspect variants you see in your filtered VCF file to become more familiar with IGV. See how quality information corresponds to alignment information at those loci. Use this website and the links therein to understand how IGV colors the alignments.