RNAseq example
Overview
Teaching: 20 min
Exercises: 30 minQuestions
How can we use experimental design in an RNAseq experiment?
Can we understand what is good and bad experimental design?
Objectives
Understand the level of replication required.
Identify whether there is enough samples to have the statistical power to answer the question.
Identify ways to improve experimental rnaseq design.
RNAseq experiment investigating the blood transcriptome
In the experiment outlined in the figure below, a group is interested in identifying differentially expressed genes (DEGs) that occur when mice are injected with an attenuated version of the causative agent of tuberculosis, Mycobacterium bovis, with a BCG injection. In order to identify DEGs they have decided on the following experimental plan.
They want to look at differences between uninfected and infected mice, so they have a control group of uninfected mice and an infected group . In each group there are two individuals. They are concerned about having replicates, but these mice are both the same strain and have been raised within a controlled environment, so they think there will not be significant variation between individuals in the same group. Instead they are going to take blood samples at 5 day intervals from day 0 to day 45 of infection. In total they will have 10 replicates from these individual mice.
The lab has a small budget for the experiment and so they are aiming to get a depth of coverage of ~20x based on the size of the mouse genome.
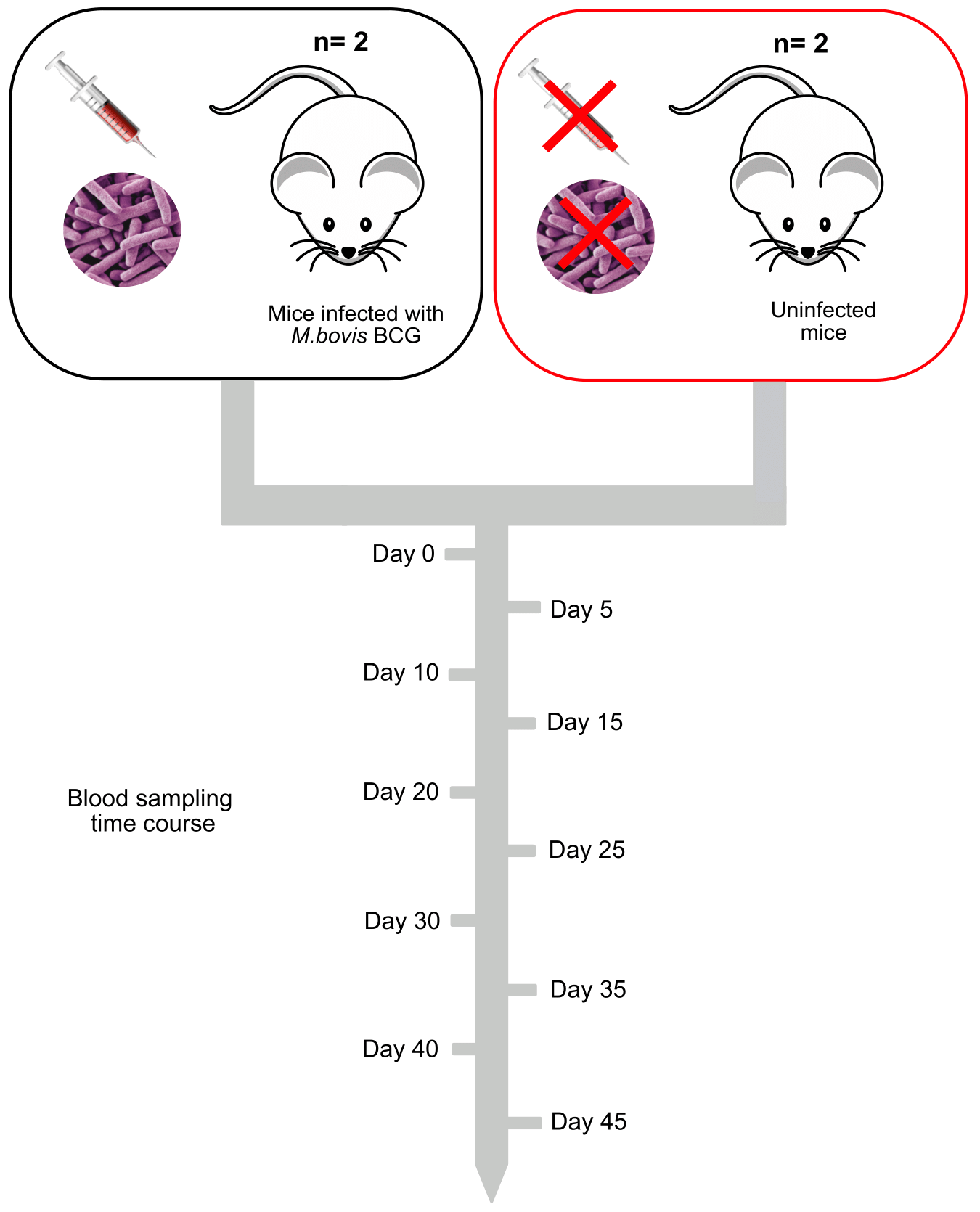
Looking at the following experimental design we are going to think about the following few questions
- How to we feel about the level of replication observed in the experimental design?
- Do we think there will be enough power in the data to identify DEGs?
- How do we feel about the depth of sequencing?
- Do we think we will be able to look at both the transcriptome over time and the comparision between uninfected and infected samples?
- How would you improve the design?
Key Points
Biological replication is much more important than technical replication in giving the experimental design power. Technical replication is still better than no replication.
We need to have appropriate controls to test hypotheses and we need to be aware of confounding variables in our designs.